Welcome to the 'Analytical Biochemistry' Group
Welcome!
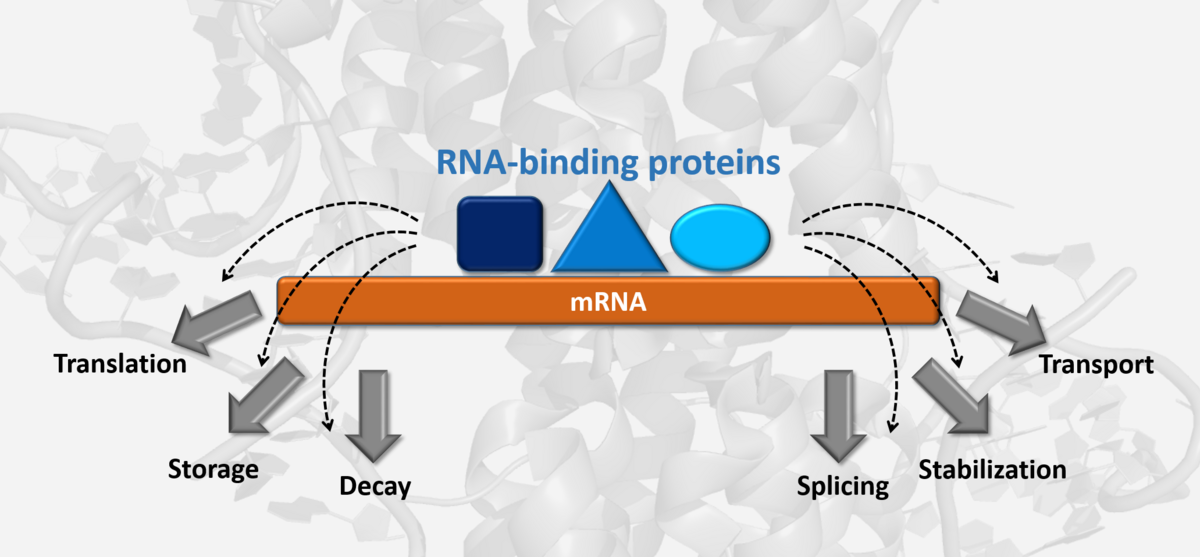
We are studying regulatory RNA-protein complexes that decide about the fate of (m)RNAs and thereby form a major regulatory parameter during cell development, immune response and cancer progression.
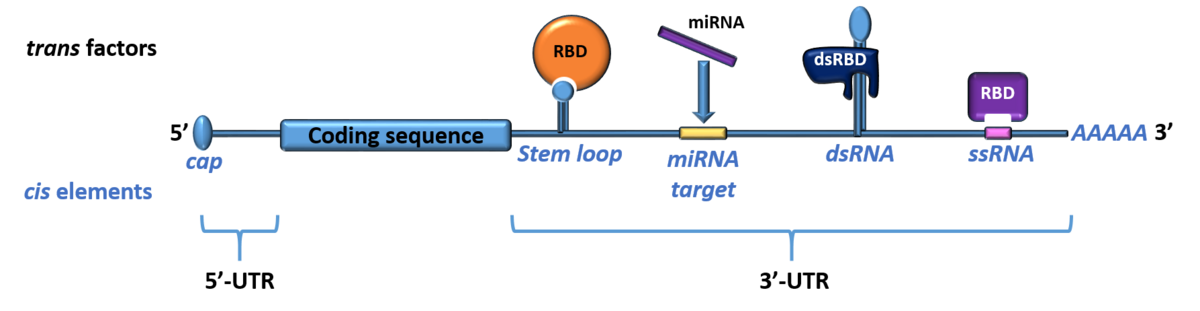
mRNAs contain untranslated regions (UTRs) in their 5’ and 3’ termini that are full of cis-regulatory elements. Those can be structured motifs, extended double-stranded stretches of RNA or short linear sequences. It appears that mRNAs are regulated through the interplay of multiple of these cis elements, often organized in hubs. trans-acting factors are mainly microRNAs and RNA-binding proteins (RBPs) with their RNA-binding domains. These domains recognize the cis elements and provoke downstream effects for the mRNA resulting e.g., in mRNA decay, protection, splicing, transport or its translational inhibition.
Apparently, the engagement of mRNA and proteins is a key checkpoint for the regulation of mRNA turnover. How nature manages to surveille specific complex formation between more than 2000 RBPs and a limited combination of the four bases has remained a secret in most of the cases. Our lab puts its efforts into unravelling rules of specificity in RNA-protein complex formation.
Here is a short summary of our activities. Details of the underlying group projects can be found by following the given links (to come...):
Specificity in RNA fold-recognition:
Nature has evolved various concepts of RNA-protein interactions. Some RBPs find their targets through a highly-specific and affine recognition of structured RNA cis elements (e.g. the by the Roquin ROQ domain). We are interested in understanding the precise RNA- and protein-based parameters for this kind of complex formation.
The specific recognition of linear RNAs:
More often, RBPs exhibit multivalent interactions through multiple RNA-binding domains (RBDs) with sequentially arranged short linear single-stranded cis elements (e.g. as known for IMP proteins). Here, the prediction and identification of the exact target mRNAs is hampered by a likely higher-order organization of extended RNA regions involved in the interaction with the protein, which is a focus of our work.
vRNPs:
Besides our interest in eukaryotic RNA-binding proteins, it appears that RNA viruses have evolved conserved RNA element structures to promote their genome and viral progression. Thus, we have developed a strong interest in the specificity of virus/virus and virus/host RNA-protein complexes.
DRBPs:
The most recent past has found a growing number of RBPs capable of interacting with DNA, and vice versa. We thus now have started to include such dual nucleic acid-binding proteins (DRBPs) into our research portfolio.
News/Jobs
Wir suchen/we are hiring: Technische Assistenz für unser neu-entstandenes Labor (feste Stelle; Eingruppierung bis E9a)!
Für weitere Infos und Bewerbung: HIER
Contact
Prof. Dr. Andreas Schlundt
Analytische Biochemie
Institut für Biochemie
Felix-Hausdorff-Straße 4
17487 Greifswald
Telefon +49 3834 420-4426
andreas.schlundtuni-greifswaldde
Achtung: Diese Seite befindet sich aktuell im Umbau!
Office:
Jana Burow
Institut für Biochemie
Felix-Hausdorff-Straße 4
17487 Greifswald
Telefon +49 3834 420-4332
jana.burow@uni-greifswald.de